Phage Discovery
In BSB 171 Biology I, students participate in the SEA-PHAGES discovery process. The semester’s goal is to accomplished something many virologists wait their entire career to achieve – identifying a new virus! Through this course centered on learning through lab work, you will become a true research scientist.

Then, in BSB 172 Biology II in the spring semester, you begin your next step entering the bioinformatics portion of the two-semester sequence. With that, you will be on your way to becoming a published research scientist. Something that few people ever become.

Our planet is inhabited by organisms at that vary widely in their levels of complexity. These organisms can be as complex as humans and as simple as a small amount of nucleic acid wrapped by a protein and lipid shell.
These various organisms interact in diverse ways that contribute to the biosphere as we know it. Among the components at the simple end of the continuum of the web of life are bacteriophage, also known as phage, which are a type of virus.
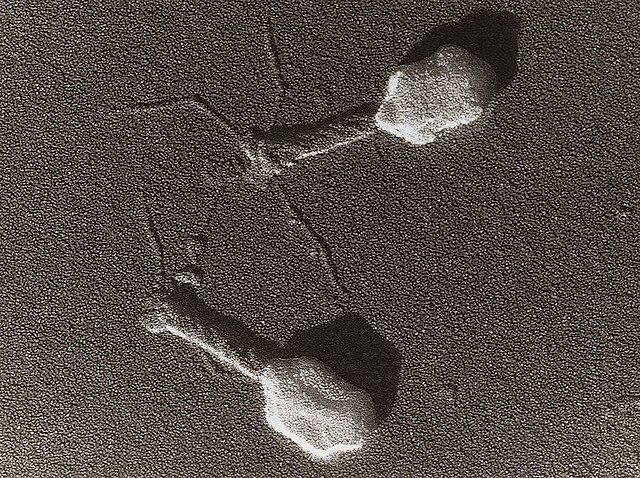
In 1915, Fredrick Twort, a British bacteriologist, identified a small agent that was able to kill bacteria. This is believed to be the first documentation of phage. Shortly after this, a French scientist at the Pasteur Institute in Paris, Félix d’Hérelle, went on to develop the clinical use of phage as antibacterial agents, today referred to as “phage therapy”. In the 1950s and 1960s, with the advent of antibiotics, phage therapy fell by the wayside.
However, with the eventual emergence of antibiotic resistant bacterial strains, there is a renewed push to develop new and novel phage therapies based on phage discoveries. In conjunction with Howard Hughes Medical Institute, one of the world’s largest basic biomedical research and science education philanthropies, we are going to follow in the footsteps of these great pioneers by isolating new and novel phage that can be characterized and potentially used for phage therapy.

In BSB 171 Biology I, the SEA-PHAGES phage discovery semester, students take environmental samples from locations around our area and isolate potentially new and novel phage that we will begin to characterize and provide the foundational knowledge required for our follow-on semester focusing on bioinformatics.

We will do this by:
- Obtaining environmental samples from the local region.
- Isolating and purifying cultures of potentially novel phage.
- Characterizing the phage isolates based on morphology, growth characteristics, host range, DNA fingerprints, and electron microscopy ultrastructure.
- Preparing isolates for archiving and genome/bioinformatic analyses.

Once we have accomplished these goals, it will culminate in the production of a presentation of our results at scientific meetings, depositing the information in phage databases, and possibly publication in a peer-reviewed scientific journal.
Continue on to:
BSB 172 Biology II – Bioinformatic analysis of phage genome phase
